White Paper
Patient Data Linkage: Maximizing Privacy, Quality, and Accuracy with Veeva SafeMine
TECHNICAL WHITE PAPER
AUTHORS & CONTRIBUTORS
Asaf Evenhaim, CEO, Veeva Crossix
Whitney Kemper, VP, Veeva Crossix Data Platform
Saar Barhoom, SVP, Research & Development, Veeva Crossix
Assaf Shtilman, Software Development Team Lead, Veeva Crossix Data Platform
Ofir Berg, Software Developer, Veeva Crossix Data Platform
Abstract
Background: As the amount of patient-level data grows rapidly, so does the complexity of linking it all. Linking patient data must be done compliantly, to preserve patient privacy, while maximizing data accuracy and completeness. There are several market solutions for privacy-preserving patient record linkage (PPRL), using various de-identification and tokenization techniques. Most industry solutions first de-identify the patient record, generate tokens for each record, and then link those records with the same token. Veeva SafeMine takes a unique approach to compliantly linking patient records over time by using the full record, including PHI, as the linking occurs behind the data supplier’s firewall leveraging federated computing and machine learning. Only then, after data has been linked with accuracy, data gets de-identified.
Objective:: The objective of this analysis is to quantify the differentiated value of Veeva SafeMine’s de-identification and tokenization technology compared to other approaches in the market. Specifically, it aims to quantify the benefit of SafeMine’s link then de-identify approach versus a de-identify then link approach.
Methods/Approach: To quantify the quality output of SafeMine’s approach relative to a de-identify then link approach, we evaluated match accuracy across two parameters: false positive and false negative matches. We performed two experiments, using two different datasets, to evaluate SafeMine’s match accuracy compared to other industry solutions. The first dataset is publicly available, synthetically generated to support privacy-preserving record linkage (PPRL) exercises. The limited sample size of this dataset is balanced by the density of data errors contained in the records. More than 80% of the identities have some data error while also including a source of truth control ID, indicating which erroneous records actually belong to the same person. Further, the synthetic data was combined into one dataset to represent and test linking multiple records for an identity from one source as one would see in health datasets such as claims, switch, pharmacy, and electronic health record (EHR), for example. Therefore, this dataset is very useful for measuring false negative matches. The second dataset is a large proprietary set of approximately 260 million identities. This dataset is representative of the U.S. adult population. This scale of identities is useful to measure the impact rate of false positive matches at scale, as it reflects identity collisions that occur in real-world large datasets.
Results: For the experiments, we determined the match accuracy of each approach based on the number of false positive and false negative matches obtained when processing the test data. A false positive indicates the engine linked records that should not be linked together. A false negative indicates the engine did not link records that should have been linked together.
For false negative matches, SafeMine’s accuracy was 87.6% versus de-identify then link’s accuracy of 34.0%. For false positive matches, SafeMine’s accuracy was 99.2% versus de-identify then link’s accuracy of 93.4%.
Conclusion: Patient data linked using SafeMine will deliver more accurate and complete linkage, supporting the integrity of patient insights and business decisions that are powered by patient data.
Introduction
With the amount of patient-level data continuing to grow exponentially, so does the complexity of linking it all. Patient data must be linked compliantly. A critical part of this is preserving patient privacy, without compromising data accuracy and completeness. This is called privacy-preserving record linkage (PPRL).
PRIVACY-PRESERVING PATIENT RECORD LINKAGE SOLUTIONS
There are several market solutions for compliant patient identity matching. These technologies use various flavors of a tokenization technique, which result in different levels of match quality. Although various methods preserve privacy, the degree of linkage quality varies significantly.
Conventional Approach
Conventional industry solutions first de-identify each patient record, by generating tokens for each record based on combinations of patient-identifying information on that record, and then attempt to combine those records with the same token.
CONVENTIONAL APPROACH
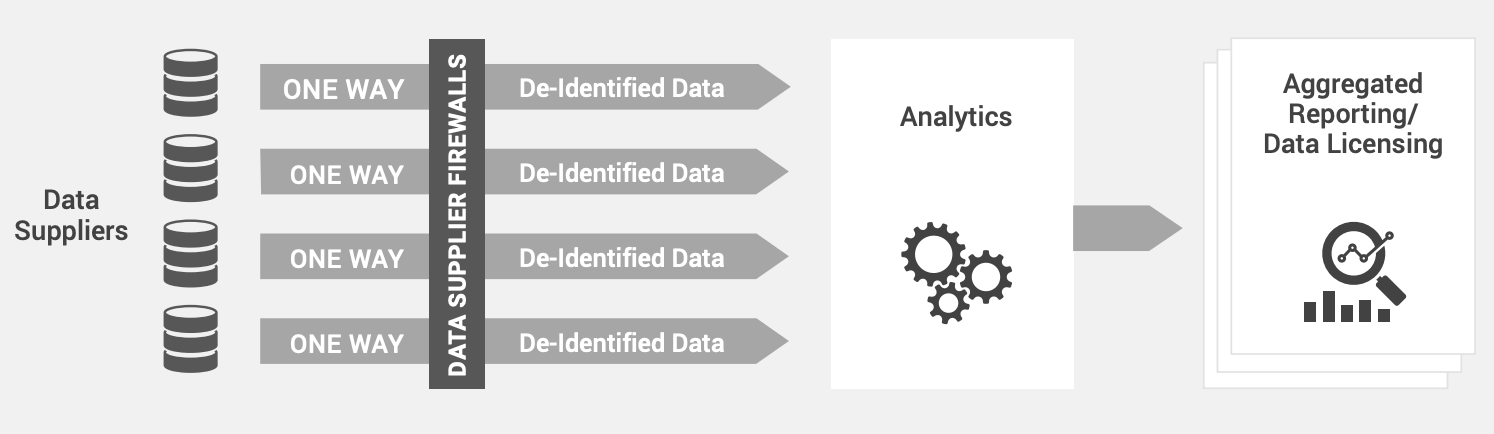
De-Identify Then Link
- Uses a deterministic match process that does not handle data entry errors well
- Not able to reference historical data for a given identity, only what is on an individual record
- Will not accurately link identities that experience life events like marriage or divorce
SafeMine Approach
Veeva SafeMine takes a unique approach to patient identity matching by linking records first and then de-identifying. SafeMine compliantly links patient records by using protected health information (PHI) and the full patient record over time, unlike the one record at a time used by conventional approaches. SafeMine linking securely occurs behind the data supplier’s firewall.
Probabilistic matching algorithms and machine-learning processes are used on identified PHI to overcome data quality issues inherent in patient data.
SAFEMINE APPROACH
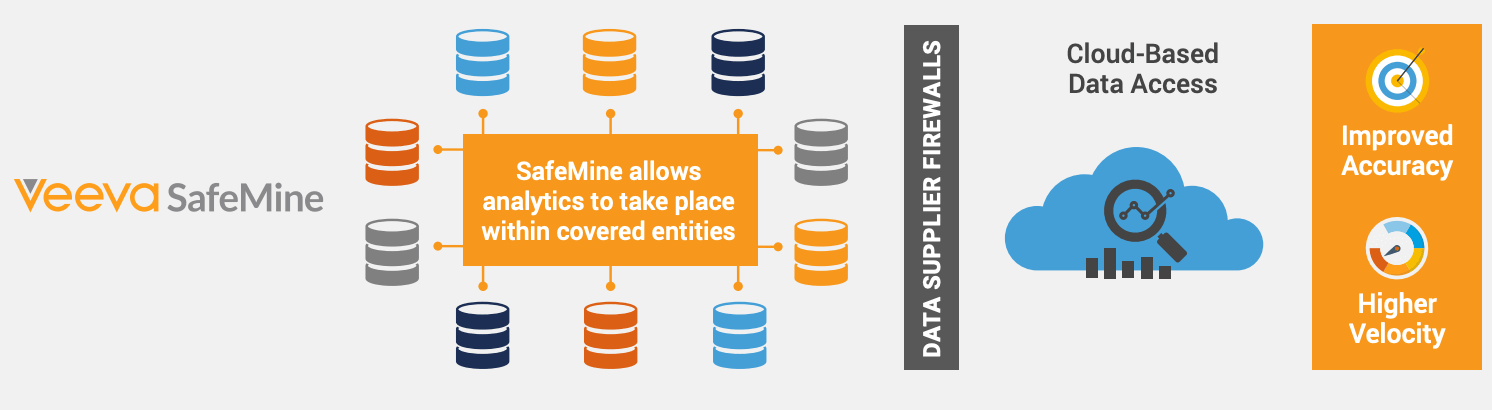
Link Then De-Identify
- Maintains a full history of PHI values associated with an identity
- Uses probabilistic match algorithms to overcome data entry errors
- Leverages historical PHI values to support AI/ML algorithms for improving match accuracy
- Monitors diversity of values for select PHI attributes to flag suspicious over-matched identities
Objective
The objective of this analysis is to quantify the differentiated value of Veeva SafeMine’s de-identification and tokenization technology compared to other approaches in the market. Specifically, it aims to quantify the benefit of SafeMine’s link then de-identify approach versus a de-identify then link approach.
Approach
To compare SafeMine’s approach to a de-identify then link approach, Veeva evaluated match accuracy across two parameters: false positive and false negative matches.

A false positive occurs when two or more records of different identities are linked, causing over-matching of records.

A false negative occurs when two or more records that belong to the same identity are not linked, causing under-matching of records.
Our evaluation included two experiments, using two different datasets
The first dataset is publicly available, synthetically generated to support PPRL1 exercises. The limited sample size of this dataset is balanced by the density of data errors contained in the records, as more than 80% of the identities have some data error.
Further, the synthetic data was combined into one dataset to represent and test linking multiple records for an identity from one source as one would see in health datasets such as claims, switch, pharmacy, and electronic health record (EHR), for example. Therefore, this dataset is very useful for measuring false negative matches.
The second dataset is a large proprietary set of nearly 260 million identities. This dataset is representative of the U.S. adult population. It is useful to measure the impact of false positive matches at scale.
Experiment 1 - Measurement of False Negative Rate (Under-match)
For the first experiment, we leveraged a publicly available data set from Synthea called Synthetic Denver.
This synthetic data is based on a population used for testing by the Childhood Obesity Data Initiative (CODI) project. It is composed of 6,357 simulated child patient records of residents in Colorado. In it, 789 identities have multiple records to test an identity matching system—more details below in Test Data Overview. This dataset represents approximately 1/100th simulation of Denver and contains realistic name, address, and contact information. The realism and variations of the demographic data is modeled after patterns found in a Denver-area healthcare provider.
The Synthetic Denver data provides a control identifier ID that is a source of truth. It establishes a unique patient identifier across the dataset. Because the expected outcome of matching identities of the dataset is clear, Veeva was able to test the match accuracy using the dataset.
The Synthetic Denver data was mapped to a flat layout so it could be processed by SafeMine and a de-identify then link engine. The data-mapping process was iterated to ensure accuracy. For example, we mapped fields such as middle name, so it was not included in the first name, as was the case in the source data.
After the mapping and data transformation was finalized, the file was processed through a series of normalizers to adjust for any formatting discrepancies or nonsensical values in the data. This process included removal of special characters, modifying date format (YYYYMMDD vs. MM/DD/YYYY), formatting zip code to five digits, and removing of future date value. The input file was then processed by SafeMine and the de-identify then link engine.
To quantify the match effectiveness of SafeMine, we processed the Synthetic Denver test data through SafeMine. We also separately processed the test data through a de-identify then link engine to determine the match effectiveness of the de-identify then link approach. We then measured and compared the false negative rates across both approaches. The match accuracy rate is based on false positive and false negative match rates.
Experiment 2 - Measurement of False Positive Rate (Over-match)
False positive rates were measured using a proprietary data set of approximately 260 million U.S. adults. Subsets of the entire dataset were processed through both a de-identify then link approach and SafeMine.
The experiment took the same random samples of 1 million, 5 million, and 20 million records from the dataset of approximately 260 million U.S. adults and processed them through both engines. The results from the experiment were then used to create a regression model that was used to project the expected values for additional sample sizes as shown in Table 4.
The experiment found the false positive rate highly dependent upon the sample size of data used, as shown in the results. Calculations were performed to measure the number of individual identities that collided, as well as the total number of collisions.
In other words, if Jane Doe collided with seven different identities, the count of identities with collisions would be one, and the total count of collisions would be seven. The false positive rate is calculated using the total count of collisions.
Furthermore, because the de-identify then link approach used two different tokens, an additional calculation was performed to determine the distinct identities between the two tokens that would result in a false positive match.
This calculation determined the union of identities between the two de-identify then link tokens that were used in the experiment, as a practical application of the tokens would need to make decisions on which token values to use. The results for distinct false positives across both tokens are shown in Table 4.
Configurations
Prior to processing the test data, SafeMine and the de-identify then link engine needed to be configured for the experiment. One of SafeMine’s strengths is the ability to configure identity matching based on the data being provided by the data supplier.
SafeMine
SafeMine uses a probabilistic match process that requires configuration based on a variety of factors.
- PHI attributes available in the source data
- This is a critical input that determines the diversity and volume of match patterns available for evaluating a match between records
- For this experiment, the following attributes were used for evaluation:
- First Name
- Last Name
- Date of Birth (DOB)
- Address
- City
- State
- Zip Code - 5 digit
- Longitude
- Latitude
- Probabilistic match algorithm
- Based on which attributes are available in the data, the algorithm will automatically select an appropriate combination of attributes to test if two records match
- Threshold values for considering a successful match or rejection
- Adjustment of these values help balance false negative and false positive rates
- Adjustments included relaxing the successful match threshold values to improve the false negative rate
De-identify then link
The de-identify then link engine uses a deterministic matching process. Therefore, the only necessary configuration was defining the match tokens. The de-identify then link engine was configured to use two token definitions for this experiment:
- Token 1 = last name + 1st initial of first name + gender + DOB
- Token 2 = last name (soundex2) + first name (soundex) + gender + DOB
These definitions were selected based on published research suggesting they provide a very high quality match rate3 and confirmation from industry stakeholders that these token definitions are most widely used across the data ecosystem.
Test Data Overview
Experiment 1 - Measurement of False Negative Rate (Under-match)
After deduplicating the source data, we identified a total of 5,478 unique patient identities. Of these identities, 789 had multiple records. Therefore, only these records can be used to test record matching. The error rate of the 789 identities was much higher than we would expect in a real-world dataset, as more than 80% of the records had a data error in a field that is used for matching.
Although the number of records is small relative to some common healthcare datasets like claims or EHR data, the diversity of data errors found in the data is representative of that seen in real-world data.
As shown in the examples below, this data provides realistic examples of common data errors that any privacy-preserving record linkage system would need to handle to accurately link the same patient identities.
The density of errors found in the test data limits the need for a high volume of data to encounter common data errors. In other words, because common data errors have been synthetically introduced into the test data, obtaining hundreds of thousands or millions of records is not necessary to encounter a variety of common data entry errors to test how well a patient matching process can handle the errors to successfully link records.
Experiment 2 - Measurement of False Positive Rate (Over-match)
SafeMine has a proprietary consumer dataset of approximately 260 million U.S. adults that was leveraged for the experiment to measure false positive rates. This dataset contains demographic information on the identities including name, address, date of birth, gender, and phone numbers. This dataset has a control identifier, assigned by a third party, that was considered a source of truth when calculating match accuracy results with this dataset.
SafeMine’s Matching Process
SafeMine’s proprietary matching process uses a series of probabilistic match algorithms to determine whether two records belong to the same identity. The two records are defined as the record to match and candidate record.
The first step in the matching process is to define the attributes that will be evaluated. For this experiment, the following attributes were used for evaluation:
- First Name
- Last Name
- Date of Birth (DOB)
- Address
- City
- State
- Zip Code - 5 digit
- Longitude
- Latitude
The next step is to determine which attributes, between the two records, can be used in SafeMine’s probabilistic match patterns. Both records must have a value for the given attribute in order for the attribute to be evaluated. The attribute value between the records is compared using a probabilistic algorithm to obtain a match score.
The probabilistic algorithm uses several techniques to ensure an accurate match. One technique is to understand the frequency of values based on the attribute. The frequency analysis goes beyond using U.S. Census data and leverages machine learning models that look across the SafeMine network to continuously optimize frequency patterns that are used to inform the match strength.
For example, SafeMine has a process to intelligently and compliantly ensure nonsensical values like ‘unknown’ or ‘N/A’ are not used in the matching process to reduce the risk of errored matches. SafeMine does this by using a machine-learning process that flags values that appear with an elevated frequency in a field so that it is ignored by the probabilistic match algorithm.
Another SafeMine technique is to use distance-based models to consider how similar two distinct values are. The distance model quantifies the similarity of the values with a score of 0 to 100, allowing configurability of tolerance levels for confirming if two identities should be considered a match. The distance model is attribute-specific and operates differently for a name field versus a date field, for example.
Based on the match score obtained from the probabilistic match process, the result for the attribute can be approved, rejected, or treated as unknown for use in the next step of the probabilistic match pattern process. An unknown result is established when the match score is between the threshold values for approved or rejected. For example, we can configure a match score of >85 to be approved, <70 to be rejected, and between 70 and 85 to be unknown. This classification is important when the final match score is calculated.
After each attribute has been scored, a list of approved, rejected, or unknown attributes is created. Based on which attributes are approved, SafeMine will dynamically generate probabilistic match patterns combining multiple attributes. The number of probabilistic match patterns is dependent upon the number and type of approved attributes. It is possible to have only a single match pattern, or more than five.
Once the match patterns are defined, they are evaluated using a probabilistic match algorithm and a match score is assigned to each pattern. Based on the score of the match pattern and the configuration of the threshold values, it is considered accepted or rejected. After each probabilistic match pattern has been scored, the algorithm will perform a final calculation using the total score of the match patterns and of the attributes to determine if the two records are a match or not. If only a single match pattern was generated, a higher match threshold value is configured.
SafeMine Monitors for False Positive Matches
SafeMine has built a robust process to identify and resolve false positive matches leveraging the longitudinal SafeMine identifier that maintains a history of the various Personally Identifiable Information (PII) attribute values associated with the SafeMine identifier. This process of learning from past decisions allows the algorithm to get better and better over time by adjusting to real data.
The process starts at each data supplier by monitoring for any over-connected identities. An overconnected identity is one that has an abnormally high diversity of values for specific PII attributes.
For example, if an identity has more than three different values for a date of birth then it is flagged as a suspicious identity for false positive matches. An identity that is flagged as suspicious for a false positive match is then processed through an identity resolution process that attempts to match the patient across attributes other than those that qualified the identity as suspicious.
For example, if the identity was flagged due to multiple date of birth values, the resolution process will attempt to identify the attribute value that caused the false positive matches. The process will look for an attribute with a common value across the over-connected records and then attempt to match the various records associated with that identity again but disabling the use of the suspected attribute that caused the over-connection.
If the records are then considered a match, the flag is removed and no other action is taken. If any records are not able to be matched (without using the suspected attribute that caused the over-matching), then a separate identity is assigned to each unresolved record, and the “offending” attribute is remembered for future matching. This process helps ensure the rate of false positives is minimized within SafeMine.
De-Identify then Link Matching Process
The de-identify then link matching engine uses a subset of attributes to create tokens for linking records as defined during configuration. Based on the token definitions for this experiment, only first name, last name, date of birth, and gender were used when generating tokens as defined above.
As a given record is processed through the de-identify then link engine, the PHI attribute values of the record, as defined for each token, get concatenated into a string. The string is then hashed using a third party key. The resulting hashed value is encrypted to create the token for the given record. If any of the PHI attribute values are missing for a given record, a token cannot be created for that record.
Each token value represents a unique identity. Therefore, when two records have the same token, they can be matched and considered as belonging to the same identity. However, since de-identify then link uses a deterministic match process, data quality errors, like misspellings of name, will result in the generation of two separate token values, preventing a successful match between records.
Results
For the experiments, we determined the match accuracy of each approach based on the number of false positive and false negative matches obtained when processing the test data by comparing the processed data to the control identifier for each dataset as the source of identity truth. A false positive indicates the engine linked records that should not be linked together. A false negative indicates the engine did not link records that should have been linked together.
Detailed match accuracy calculation is detailed below:
- A unique identity generated by SafeMine engine = SafeMine Patient Identifier (PID)
- A unique identity generated in de-identify then link engine = Token
- False positive rate = # of PIDs or Tokens with multiple control identifiers / Total # of control identifiers
- False negative rate = # of control identifiers with multiple PIDs or Tokens / Total # of control identifier
Table 1: Experiment 1 - False Negative Match Accuracy Results
In an extremely messy dataset, with more than 80% of records having an error, SafeMine significantly outperformed the de-identify then link engine across (1) those records with multiple entries and (2) those with a single entry.
In the population with multiple entries, which allows for the measure of longitudinal linking quality across multiple sites of care, SafeMine performed 5 times better than de-identify then link engine when normalizing the data error rate to the industry average of 45%.
| Count | % using 789 with multiple entries | % using all 5478 patients | |
|---|---|---|---|
| False Negative | 98 | 12.42% | 1.79% |
| Count | % using 789 with multiple entries | % using all 5478 patients | |
|---|---|---|---|
| Token 1 - False Negative | 526 | 66.67% | 9.60% |
| Token 2 - False Negative | 515 | 65.27% | 9.40% |
Table 2: Experiment 2 - False Positive Match Accuracy Results
SafeMine outperformed the de-identify then link approach across all sample sizes and most notably at the largest sample sizes with SafeMine’s rate being 8.5x better than de-identify then link. At small sample sizes, like 5M identities, the rates of false positives for both approaches were very good at around 1% with SafeMine and demonstrating 5x fewer false positive results as compared to the union of the two de-identify then link tokens, 13,294 vs. 70,143.
Table 2 details some examples of where SafeMine was able to successfully reject a match that the de-identify then link approach inaccurately matched.
| Sample Size | Count of Identities with Collisions | % of Identities with Collisions | Total Count of Collisions | False Positive Rate |
|---|---|---|---|---|
| 1,000,000 | 1,211 | 0.12% | 2,427 | 0.24% |
| 5,000,000 | 6,636 | 0.13% | 13,294 | 0.27% |
| 10,000,000 | 13,697 | 0.14% | 27,795 | 0.28% |
| 20,000,000 | 29,366 | 0.15% | 58,860 | 0.29% |
| 50,000,000 | 84,897 | 0.17% | 179,795 | 0.36% |
| 100,000,000 | 209,897 | 0.21% | 459,795 | 0.46% |
| 150,000,000 | 374,897 | 0.25% | 839,795 | 0.56% |
| 200,000,000 | 579,897 | 0.29% | 1,319,795 | 0.66% |
| 250,000,000 | 824,897 | 0.33% | 1,899,795 | 0.76% |
| 259,000,000 | 873,245 | 0.34% | 2,014,815 | 0.78% |
| Sample Size | Count of Identities with Collisions | % of Identities with Collisions | Total Count of Collisions | False Positive Rate |
|---|---|---|---|---|
| 1,000,000 | 2,920 | 0.29% | 5,886 | 0.59% |
| 5,000,000 | 34,296 | 0.69% | 70,143 | 1.40% |
| 10,000,000 | 77,643 | 0.78% | 157919 | 1.58% |
| 20,000,000 | 184,298 | 0.92% | 373,621 | 1.87% |
| 50,000,000 | 605,643 | 1.21% | 1,225,919 | 2.45% |
| 100,000,000 | 1,715,643 | 1.72% | 3,460,919 | 3.46% |
| 150,000,000 | 3,325,643 | 2.22% | 6,695,919 | 4.46% |
| 200,000,000 | 5,435,643 | 2.72% | 10,930,919 | 5.47% |
| 250,000,000 | 8,045,643 | 3.22% | 16,165,919 | 6.47% |
| 259,000,000 | 8,568,543 | 3.31% | 17,214,419 | 6.65% |
*Bold values represent actual result, all others are projected
Examples of Benefits of Probabilistic Matching over Deterministic Matching
Table 3 - False Negative Example Records and Match Results
This table provides specific examples of how SafeMine was able to successfully link records that the de-identify then link engine considered two separate identities. SafeMine is able to successfully link records even when data errors exist by leveraging its probabilistic match algorithm.
Each letter in the last three columns represents a unique token value for each token type. Green indicates correct match results. Red indicates incorrect match result.
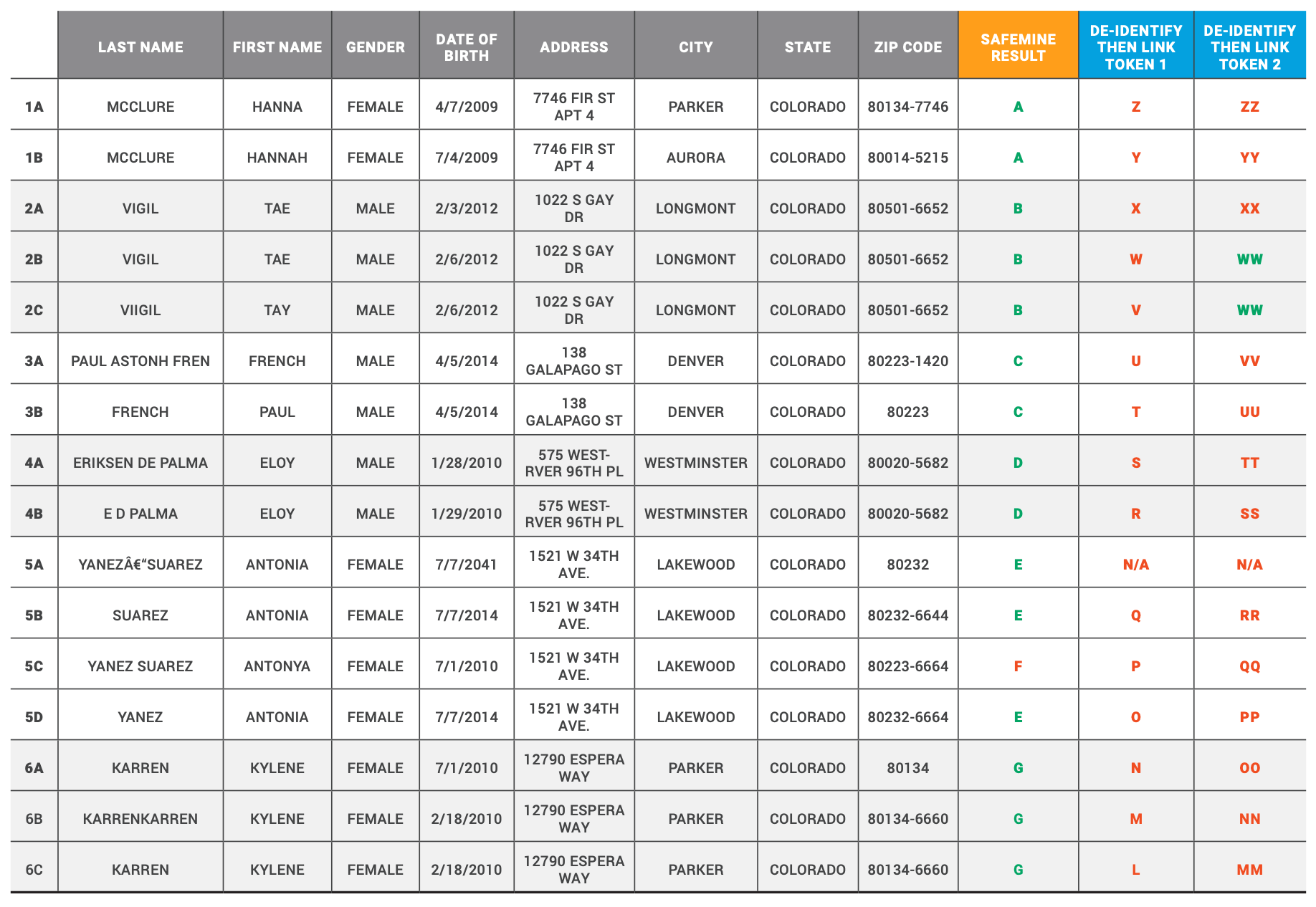
1. DATE OF BIRTH MONTH AND DAY VALUES SWAPPED AND FIRST NAME ERRORS
The identity in Table 3, example 1 has two records, 1a and 1b. The records have different dates of birth, a small difference in the spelling of the first name, different city names, and different zip codes. Note, the upper/lower case differences in the last name do not impact the match process as the normalizers convert the strings to all capital letters.
The de-identify then link engine generated a different token for each record. The different token values can be attributed to the different dates of birth found in the records as the first name variations would be addressed in Token 1 by only taking the first initial and Token 2 by the soundex process. However, neither token could account for the data-entry error of switching the date of birth day and month.
SafeMine leveraged its probabilistic matching process to generate the same SafeMine identity value for both records. The probabilistic matching process was able to overcome the numerous data-entry errors by not only leveraging the PHI attributes between the records that are consistent, like last name and address, but also recognizing that the date of birth values are similar to establish a match between the records.
2. DATE OF BIRTH AND LAST NAME DIFFERENCES
The identity in Table 3, example 2 has three records, 2a, 2b, and 2c. Record 2a has a date of birth that is different from 2b and 2c. Record 2c has a last name and first name that is slightly different from 2a and 2b.
The de-identify then link engine created different values for Token 1 and Token 2 for records 2a and 2b due to different date of birth values. However, for record 2c, only Token 1 generated a unique value whereas Token 2 replicated the token value from record 2b. The reason Token 2 was able to account for the extra ‘i’ entered in the last name and the slight difference in spelling of the first name is its use of soundex for the name fields. In this example, the soundex methodology was able to account for the slight variations in name fields.
SafeMine’s probabilistic matching process was able to account for the different date of birth values and the slight differences in the name fields to successfully create the same identity value across all three records 2a, 2b, and 2c. First, record 2a and 2b were compared and matched successfully as the probabilistic match algorithm recognized a slight difference in date of birth but was able to use other fields to confirm they belong to the same identity.
Then SafeMine evaluated records 2a and 2c. Again a match was confirmed as the probabilistic match algorithm recognized that the first name, last name, and date of birth only had a slight variation. Further, the fields without a difference—address, city, state, zip code—resulted in a high enough score to confirm the match.
Finally, SafeMine evaluated records 2b and 2c. Again a match was confirmed as only the name fields had a slight variation that the probabilistic match algorithm was able to overcome to confirm the match.
3. FIRST NAME AND LAST NAME VALUES ARE SWAPPED
The identity in Table 3, example 3 has two associated records with data-entry errors with record 3a swapping the first name and last name, and the last name value contains the first, middle, and part of the last name of the identity.
The de-identify then link engine created two separate tokens for Token 1 and Token 2, as the first and last name fields are significantly different. Therefore, neither taking only the first initial of the first name nor applying soundex are able to overcome the data-entry errors.
SafeMine recognized that the first and last name values are not a match and excluded those values from its probabilistic match process. Instead, SafeMine was able to use date of birth, address, and zip to confirm that records 3a and 3b belong to the same identity.
4. LAST NAME ABBREVIATED AND DOB ERROR
The identity in example 4 has two associated records with record 4a having the last name fully spelled out and 4b abbreviating the last name. Also, the date of birth values differ by one day between the records.
The de-identify then link engine created two separate tokens for Token 1 and Token 2 as the slight difference in date of birth will result in different values for both definitions. Even if the date of birth difference did not exist, the different values in last name would have resulted in different Token 1 and Token 2 values. For Token 1, the different values would be due to the different string values in the last name. For Token 2, the different values in last name result in different soundex values. For example EriksenDePalma is converted to E625 and EDPalma is converted to E314. Therefore, Token 2 would generate two different values for records 4a and 4b even if the date of birth values were the same.
SafeMine successfully accounted for the difference in last name and date of birth through the use of its probabilistic match process. SafeMine recognized the slight variations in date of birth and last name. SafeMine assigned an unknown value regarding the match and not a rejection due to the probabilistic match algorithm recognizing that the values are similar. Therefore, with the other field values being consistent, SafeMine was able to successfully match the two records.
5. ZIPCODE, FULL NAME, AND DOB ERRORS
The identity in example 5 has four associated records and a variety of differences between them. Record 5a has special characters in the last name and a future-dated year of birth as the last two digits appear to be swapped. Note, the normalizer would remove the special characters and the future-dated date of birth value. Record 5b only has one value in the last name as compared to 5a and 5c, which have two values. Record 5c has a different spelling for the first name and a different date of birth month and year as compared to the other three records. Record 5d has only one value in the last name field, and that value is different from the single value found in record 5b.
The de-identify then link approach created three separate token values for Token 1 and Token 2 for records 5b, 5c, and 5d. For record 5a, no token value was generated for Token 1 or Token 2 due to the normalizer removing the date of birth value, which represented a date in the future. Records 5b and 5c were different for Token 1 and Token 2 due to different dates of birth. Records 5b and 5d were different for Token 1 and Token 2 due to different last name values. Records 5c and 5d were different for Token 1 and Token 2 due to different dates of birth.
SafeMine was able to successfully match records 5a, 5b, and 5d. However, given the differences in last name, first name, and date of birth, record 5c was assigned a different SafeMine ID as compared to the other records associated with this identity. Records 5a and 5b have different last names and a missing value in date of birth, so no match can be evaluated on that attribute. However, first name, address, city, state, zip, gender, and longitude/latitude were confirmed to match. The probabilistic match algorithm was able to establish these two records as a match and assign the same SafeMine identity value.
Records 5a and 5c were not matched successfully. SafeMine considered the last name and first name values as an unknown match; date of birth was not evaluated due to missing value in record 5a; zip code was rejected as a match; and address city, state, and longitude/latitude were considered a match. When the probabilistic match patterns across the matched and unknown attributes were evaluated and scored, the result was not significant enough to confirm a match. Therefore, SafeMine rejected the match between these records.
SafeMine was able to successfully match records 5a and 5d. Because the last name of record 5a was cleansed by the normalizer process, SafeMine found that all other attribute values matched between the records and confirmed the match by assigning the same SafeMine identifier to both records.
SafeMine was able to successfully match records 5b and 5c. SafeMine considered the first name values as an unknown match; the zip code values as not a match; last name was confirmed as a match despite the differences; and the other attribute values were considered a match. The matched and unknown attribute values were used in the probabilistic match process and the match score was significant enough to establish a match between the records. Therefore, record 5c has one match accepted (5b) and one match rejected (5a). There are more records to evaluate before the final result is determined.
SafeMine was able to successfully match records 5b and 5d. Only the last name values were considered not a match. Address, city, state, zip code, longitude/latitude, first name, and date of birth were calculated to be a match. The probabilistic match patterns established these two records as a match.
Finally, SafeMine was not able to successfully match records 5c and 5d. SafeMine considered the last name and first name values as unknown matches; date of birth and zip code as not a match; and address, city, state, and longitude/latitude as a match. Based on these results, the probabilistic match process rejected a match between these records. Therefore, record 5c has one comparison that had a successful match (5b) and two comparisons that were not successful (5a and 5d). Therefore, the final result for 5c is the creation of a SafeMine identifier that is different from 5a, 5b, and 5d.
The result of the matching process across these four records is the creation of two different SafeMine identities. The first identity successfully linked the records in rows 1, 2, and 4. However, row 3 was not linked and was assigned its own SafeMine identity value. Therefore, this is considered a false negative score for SafeMine as all four records were not successfully matched.
6. LAST NAME ENTERED TWICE AND DOB ERRORS
The identity in example 6 has three records, with record 6a having a date of birth that is different from 6b and 6c, and record 6b having a last name value that duplicated the value found in 6a and 6c.
The de-identify then link approach created three different token values for Token 1 and Token 2. Record 6a has a different date of birth from 6b and 6c, so both Token 1 and Token 2 will assign a unique token value to the record. Record 6b has a different last name as compared to 6a and 6c, so both Token 1 and Token 2 will assign a unique token value to the record. Given the results for 6a and 6b, record 6c will have a unique token value for both Token 1 and Token 2. This example provides another use case where soundex is not able to overcome a data-entry error in the name field. The soundex value for KarrenKarren is K652 and the soundex value for Karren is K650. Therefore, Token 2 is not able to confirm that records 6b and 6c belong to the same identity.
SafeMine successfully linked the three records using a probabilistic match process to overcome the data-entry errors found in the date of birth and last name fields. For records 6a and 6b, the date of birth was rejected, last name was considered unknown, and the match was confirmed using the probabilistic match process with all fields except date of birth. For records 6a and 6c, only the date of birth was rejected, so all other attributes were used by the probabilistic match process to confirm the match. Finally, for records 6b and 6c, the date of birth was rejected, last name was unknown, and the remaining attribute values were used by the probabilistic match process to confirm the match. Therefore, SafeMine was able to successfully link the three records in this example.
Table 4 - Examples of False Positive Results by De-Identify Then Link
This table provides specific examples of how SafeMine was able to successfully assign tokens to individual records that the de-identify then link engine incorrectly considered to be the same identity. SafeMine is able to successfully assign unique tokens to records by leveraging its probabilistic match algorithm.
Each letter in the last three columns represents a unique token value for each token type. Green indicates correct match results. Red indicates incorrect match result.
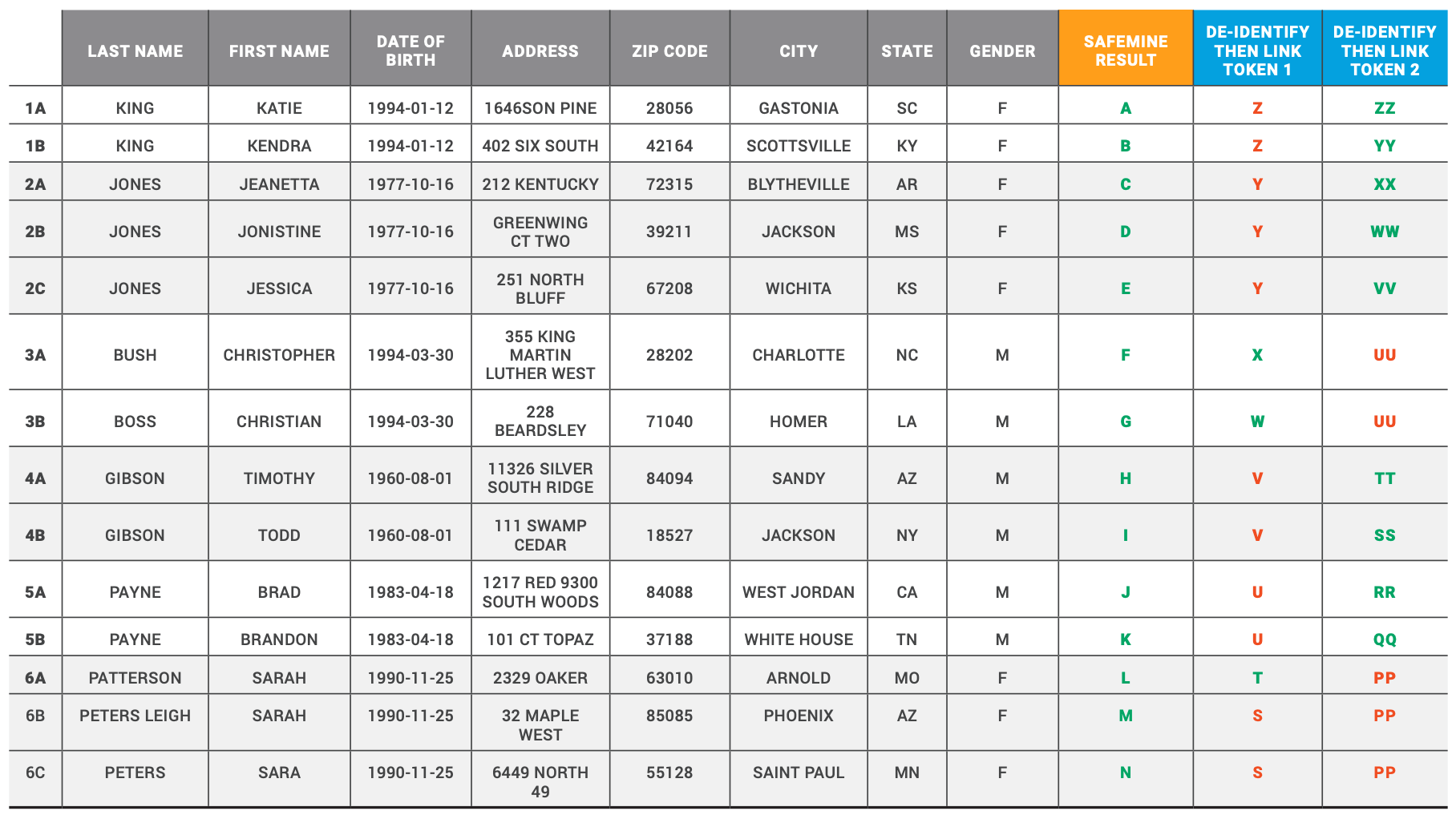
1. DIFFERENT FIRST NAME AND ADDRESS
In Table 4, example 1, there are two records that were incorrectly linked by de-identify then link Token 1 only. SafeMine and de-identify then link Token 2 were able to use their match processes to reject the linking of records 1a and 1b.
Token 1 was not able to determine the difference between the full first name and addresses associated with these records, so it inaccurately assigned the same token values to record 1a and 1b.
SafeMine’s match algorithm was able to find enough of a difference in the first name and address fields to ensure records 1a and 1b were not merged. Similarly, de-identify then link token 2 was able to leverage the soundex value for the different first names to ensure different token values were created for de-identify then link Token 2.
2. MULTIPLE DIFFERENT FIRST NAMES AND ADDRESSES
In Table 4, example 2 there are three records that were incorrectly linked by de-identify then link Token 1 only. SafeMine and de-identify then link Token 2 were able to use their match processes to reject the linking of records 2a, 2b, and 2c.
By only using the first initial of the first name, de-identify then link Token 1 was not able to determine the differences between the full first name and addresses associated with these records, so it inaccurately assigned the same token values to all three records.
SafeMine’s match algorithm was able to find enough of a difference in the first name and address fields to ensure the three records were not merged. Similarly, de-identify then link token 2 was able to leverage the soundex value for the different first names to ensure different token values were created for de-identify then link Token 2.
3. DIFFERENT FIRST NAME, LAST NAME, AND ADDRESS
In Table 4, example 3, there are two records that were incorrectly linked by de-identify then link token 2 only. SafeMine and de-identify then link Token 1 were able to use their match processes to reject the linking of records 3a and 3b
The reliance on de-identify then link Token 2 on soundex caused an incorrect match in this example as the soundex values for the last name and first name between these two records results in the same value, B200 and C623, respectively. Therefore, de-identify then link token 2 created the same token values for both records.
SafeMine and de-identify then link Token 1 were able to recognize the differences in name and address to create two separate identities for the records in this example.
4. DIFFERENT FIRST NAME AND ADDRESS
In Table 4, example 4, there are two records that were incorrectly linked by de-identify then link Token 1 only. SafeMine and de-identify then link token 2 were able to use their match processes to reject the linking of records 4a and 4b.
Token 1 was not able to determine the difference between the full first name and addresses associated with these records, so it inaccurately assigned the same token values to record 4a and 4b.
SafeMine’s match algorithm was able to find enough of a difference in the first name and address fields to ensure records 4a and 4b were not merged. Similarly, de-identify then link token 2 was able to leverage the soundex value for the different first names to ensure different token values were created for de-identify then link Token 2.
5. DIFFERENT FIRST NAME AND ADDRESS
In Table 4, example 5, there are two records that were incorrectly linked by de-identify then link Token 1 only. SafeMine and de-identify then link Token 2 were able to use their match processes to reject the linking of records 5a and 5b.
Token 1 was not able to determine the difference between the full first name and addresses associated with these records so it inaccurately assigned the same token values to record 4a and 4b.
SafeMine’s match algorithm was able to find enough of a difference in the first name and address fields to ensure records 5a and 5b were not merged. Similarly, de-identify then link token 2 was able to leverage the soundex value for the different first names to ensure different token values were created for de-identify then link token 2.
6. MULTIPLE DIFFERENT FIRST NAMES, LAST NAMES AND ADDRESSES
In Table 4, example 6, there are three records that were incorrectly linked by de-identify then link token 2 only. SafeMine and de-identify then link token 1 match processes were able to account for different name values and address values across the three records to create separate tokens for each record, 6a, 6b, and 6c. De-identify then link token 1 created separate token values for each record as the last name values were different enough to generate different token values.
SafeMine was able to use the differences in last name, first name, and address to ensure the three records, in this example were not incorrectly linked.
However, de-identify then link token 2 created the same token for all three records as the soundex values for the different first name (S600) and last name (P362) values are all the same. This example highlights some of the challenges with using the soundex algorithm for linking identities.
Conclusion
SafeMine’s probabilistic match algorithm successfully differentiates between data-entry errors and truly different values to successfully confirm which records belong to the same identity significantly more accurately than a deterministic approach.
The numerous examples expose the weaknesses of only using a de-identify and then link approach to linking patient records. Being able to perform probabilistic match analysis on the actual values in the source data also increases the efficacy of match results. Further, SafeMine’s ability to maintain historical records for an identity and use machine learning processes to link data using PHI values before de-identifying increases the robustness of its match process.
Relying on a de-identify and then link approach will provide a lower quality of patient matching as slight differences in last name, first name, gender and date of birth will result in unsuccessful matches. Further, this approach struggles with accounting for life events like marriage and divorce, casting doubt or leading to inaccurate insights when the data is used in analytics. When poorly matched patient data is used to generate actionable insights, the actions taken can be inefficient and/or inaccurate, reducing credibility and wasting resources.
SafeMine’s probabilistic match algorithm is the only market solution that is accurate and robust enough to handle the rapidly growing volume of patient-level data in the U.S., and minimize both false positive and false negative matches at scale.
SafeMine delivers a superior approach to privacy-preserving patient record linkage. Patient data linked using SafeMine will deliver more accurate and complete insights, and help optimize the business decisions that are powered by patient data.
Appendix
Limitations of Soundex for Matching Names
An article published by Frankie Patman and Leonard Shaefer from Language Analysis Systems, Incorporated, captures the limitations of using soundex algorithms for linking names.4
An excerpt from the article details specific use cases that soundex struggles with. See article for detailed examples.
- Dependence on initial letter
- Noise intolerance
- Differing transcription systems
- Names containing particles
- Perceptual differences
- Silent consonants
- Name syntax variation
- Name equivalence
- Use of Initials
- Unranked, unordered returns
- Poor precision
Despite the widespread use of soundex as an algorithm to improve match accuracy between names, it leaves much to be desired relative to a more robust probabilistic matching algorithm like that found in SafeMine
1https://synthea.mitre.org/downloads
2https://www.archives.gov/research/census/soundex
3http://careersdocbox.com/Nursing/74191628-Matching-accuracy-of-patient-tokens-in-de-identified-health-data-sets.html
4chrome-extension://efaidnbmnnnibpcajpcglclefindmkaj/viewer.html?pdfurl=https%3A%2F%2Fwww.immagic.com%2FeLibrary%2 FARCHIVES%2FGENERAL%2FLAS_US%2FL030206B.pdf&clen=665920&chunk=true